Mohammad Jobayerh
Department of Microbiology, National Center for Control of Rheumatic Fever & Heart Disease, Dhaka, Bangladesh.
Mizanur Rahman
Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh.
Nadira Akter
Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh.
Naomee Shareef
Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh.
Rubina Afroz Rana
Department of Microbiology, National Center for Control of Rheumatic Fever & Heart Disease, Dhaka, Bangladesh.
SM Shamsuzzaman
Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh.
Keywords: Antibiotic susceptibility, Pus, Resistance, Wound swab.
DOI: 10.3329/bmrcb.v47i2.57777
Abstract
Background: Antimicrobial resistance is a global health problem and the number of organisms developing resistance to commonly used antibiotics is increasing.
Objective: The study was aimed to find out the pattern of common organism isolated from wound swabs and pus with their antibiogram.
Methods: This observational study was conducted from September 2018 to January 2019 in a tertiary care hospital in Dhaka, Bangladesh. Culture and sensitivity tests were done for wound swabs and pus samples. Data regarding information of the patients, isolated organisms and culture and sensitivity reports were collected from the records of the department of microbiology.
Results: Out of 1709 samples, 72.0% yielded growth of organisms of which 86.4% were gram negative and 13.6% were gram positive bacteria. Pseudomonas spp was the most commonly (43.8%) isolated organism from both wound swab and pus samples followed by Escherichia coli (16.6%), Staphylococcus aureus (11.8%), Klebsiella spp (9.8%). Among gram negative bacteria, 14.9% were ESBL producing organisms and Klebsiella spp were the most commonly isolated ESBL producers. Most of the bacteria showed high resistance to commonly used antibiotics. Gram negative bacteria were mostly resistant to amoxicillin followed by fluoroquinolones, co-trimoxazole, and cephalosporins whereas colistin, carbapenems and piperacillin/tazobactum were the most effective drugs against them. Majority of gram positive bacteria were resistant to fluoroquinolones and co-trimoxazole but 100% Staphylococcus aureus were sensitive to vancomycin, followed by linezolid (98.0%) and teicoplanin (86.0%) and 32.0% of them were Methicillin resistant (MRSA).
Conclusion: The susceptibility pattern shows that some common antibiotics, especially antibiotics of oral form have very limited usefulness in treatment of infections and also highlight the need for regular reporting and antibiogram guided antibiotic prescription.
Keywords: Antibiotic susceptibility, Pus, Resistance, Wound swab.
Introduction
Wound infection is one of the most common hospital acquired infections and is an important cause of morbidity and mortality worldwide.1 Postoperative wound infection delays recovery, increased hospital stay and may produce long lasting sequelae.2 Skin and soft tissue infections are caused by microbial pathogens after trauma, burn or surgical procedures result in production of pus.3,4 Bacterial infection is a serious problem to the successful treatment of wound resulting in complications sometimes leading to fatal sepsis.5
Spectrum of isolated pathogens usually depends on surgical procedure involved, hospital environment and also the infection prevention policy of the hospital.6 Prevalent organisms associated with wound infection and pus formation include Staphylococcus aureus, Enterobacteriaceae, Pseudomonas aeruginosa, Coagulase Negative Staphylococci (CoNS), Enterococci.7,8
Use of antimicrobial agents cause a ‘selective pressure’ on microbial population.9 As a result of indiscriminate use of antimicrobial agents, significant changes occur in microbial genetic ecology.10 The increasing frequency of antimicrobial resistance among pathogens causing nosocomial and community acquired infections is making numerous classes of antimicrobial agents less effective.11
The emergence of antibiotic resistance and its rapid spread among pathogenic bacteria is a grave threat to public health worldwide. During last few decades, multidrug-resistant bacterial strains such as Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Methicillin resistant Staphylococcus aureus (MRSA) are increasingly associated with infections under hospital settings.12,13 Extended-spectrum beta-lactamases (ESBLs) are enzymes that mediate resistance to extended spectrum e.g., third generation cephalosporins and the incidence of ESBLs in gram negative bacteria has increased in recent years.14,15
An in-depth knowledge of the pattern of predominant organisms in pus and wound is essential for the treatment of the patient before getting the result of microbiological culture. This would be crucial to reduce the overall infection related morbidity and mortality. The present study was therefore designed to determine the current microbial spectrum of wound infection with their antibiotic sensitivity pattern.
Materials and Methods
This observational study was conducted in the Department of Microbiology of Dhaka Medical College, Dhaka, Bangladesh for a period of five months from September 2018 to January 2019.
All wound swabs and pus samples from both inpatient and outpatient departments of Dhaka Medical College Hospital sent to the microbiology laboratory for culture and sensitivity test regardless of age, sex and antibiotic intake were included in this study. Data regarding the identity of the patient, referring departments, type of specimen and sensitivity reports were collected from the records of the laboratory.
Microbiological methods
Culture of wound swab and pus: Samples were inoculated in blood agar and MacConkey’s agar media and incubated aerobically at 37ºC for 24 hours.
Isolation and identification of bacteria: The inoculated plates were examined for bacterial growth and organisms were identified by colony morphology, hemolytic criteria, pigment production, Gram stain and different biochemical tests like catalase test, coagulase test, oxidase test, and reaction in TSI agar, MIU and Simmon’s citrate agar media.
Antimicrobial susceptibility test: Antimicrobial susceptibility pattern of isolated organisms were done following Kirby-Bauer disc diffusionmethod.16 Sensitivity was done using commercially available antibi•otic discs (Oxoid, UK); amikacin (30ìg), amoxyclav (20ìg amoxycillin/10ìg clavulanic acid), cefoxitin (30ìg), ceftazidime (30ìg), ceftriaxone (30ìg), clindamycin (2ìg), ciprofloxacin (5ìg), colistin (10ìg), doxycycline (5ìg), gentamicin (10ìg), imipenem (10ìg), levofloxacin (5ìg), linezolid (30ìg), meropenem (10ìg), piperacillin/tazobactum (100/10ìg), teicoplanin (30ìg), vancomycin (30ìg). Zone of inhibition was measured according to the CLSI guideline.14
Detection of ESBL: ESBL production in gram negative bacteria was detected by double disc synergy test following CLSI guideline using amoxyclav and 3rd generation cephalosporins discs.14
Data management: Collected data were classified according to characteristics and various statistical methods and ‘Microsoft Excel’ software were used for analysis.
Results
A total of 1709 samples (wound swabs and pus) were cultured. Of them 1083 were wound swabs; among which 846 (78.1%) yielded growth of bacteria and 626 samples were pus of which 385 (61.5%) were positive in culture. Difference between the isolation rate of organisms from wound swab and pus is statistically significant (p<.001, Chi-Squire test, df=1; 95% CI). A total 1231 (72.0%) samples yielded growth of bacteria (table I).
Samples |
Growth (%) |
No Growth (%) |
Total (%) |
P value |
|---|---|---|---|---|
Wound swab |
846 (78.12) |
237 (21.88) |
1083 (100) |
<.0001s |
Pus |
385 (61.50) |
241 (38.50) |
626 (100) |
<.0001s |
Total |
1231 (72.03) |
478 (27.97) |
1709 (100) |
|
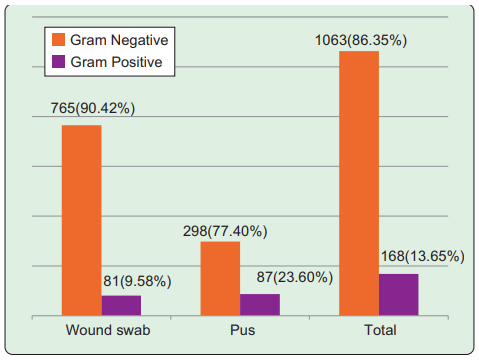
Among total 1231 culture positive samples 1063 (86.4%) yielded growth of gram negative bacteria and 168 (13.7%) had growth of gram positive bacteria. In wound swabs, 765 (90.4%) were gram negative and 81 (9.6%) were gram positive bacteria; in pus samples, 298 (77.4%) had growth of gram negative and 87 (23.6%) were gram positive bacteria (Figure 1)
Pseudomonas spp was the most commonly isolated organism (43.8%) from both wound swab and pus samples. Escherichia coli was isolated from 16.6% samples, 9.8% samples yielded growth of Klebsiella spp and Proteus spp was isolated from 7.6% samples. Staphylococcus aureus was the most frequently isolated (11.8%) gram positive bacteria from both wound swab and pus (table II).
Organism |
Wound swab Isolates n (%) |
Pus Isolates n (%) |
Total Isolates n (%) |
|---|---|---|---|
Pseudomonasspp |
431 (50.95) |
108 (28.05) |
539 (43.79) |
E. coli |
102 (12.06) |
102 (26.49) |
204 (16.57) |
Stah aureus |
68 (8.04) |
77 (20.00) |
145 (11.78) |
Klebsiella spp |
89 (10.52) |
32 (8.31) |
121 (9.83) |
Proteusspp |
73 (8.63) |
20 (5.19) |
93 (7.55) |
Enterobacterspp |
44 (5.20) |
21 (5.45) |
65 (5.28) |
Acinetobacter spp |
13 (1.54) |
7 (1.82) |
20 (1.62) |
Citrobacterspp |
8 (0.95) |
6 (1.56) |
14 (1.14) |
CoNS |
6 (0.71) |
7 (1.82) |
13 (1.06) |
Streptococcous |
5 (0.59) |
3 (0.78) |
8 (0.65) |
Serratiaspp |
5 (0.59) |
2 (0.52) |
7 (0.57) |
Enterococci spp |
2 (0.24) |
0 (0.00) |
2 (0.16) |
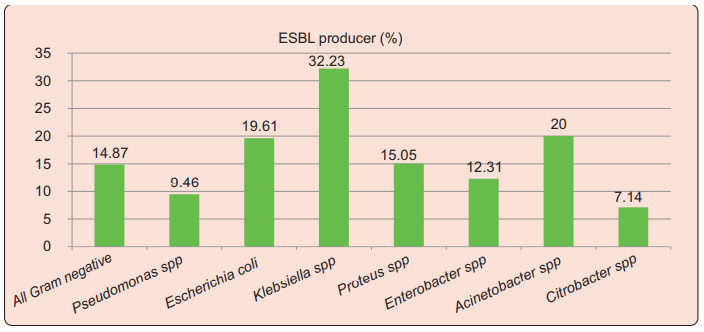
Among the isolated gram negative bacteria, 14.7% (157/1056) were ESBL producing organisms. Klebsiella spp were the most commonly isolated ESBL producers, 32.2% of which produced ESBLs. Among Pseudomonas spp 9.5% were ESBLs producer; 19.61% Escherichia coli, 15% of Proteus spp, 12.3% Enterobacter spp and 20% of isolated Acinetobacter spp were ESBL producing organisms (Figure 2).
Carbapenems were very effective antibiotics against
E. coli, Klebsiella spp. Proteus and Enterobacter spp; colistin was found most effective antibiotic in vitro against all gram negative except Proteus spp. Piperacillin/tazobactum was a very effective antibiotic in vitro against Enterobacter, Proteus and Citrobacter spp (table III).
Organism |
Sensitivity (%) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Ak |
Aml |
Amc |
Caz |
Cro |
Cip |
Cot |
L |
G |
Imp |
Mem |
Tzp |
CT |
|
Pseudomonas |
27 |
- |
8 |
56 |
9 |
20 |
22 |
11 |
27 |
66 |
68 |
62 |
97 |
E. coli |
43 |
7 |
10 |
20 |
23 |
21 |
19 |
39 |
41 |
91 |
82 |
71 |
95 |
Klebsiella |
48 |
- |
9 |
11 |
13 |
18 |
29 |
48 |
30 |
87 |
85 |
65 |
93 |
Proteus |
33 |
3 |
13 |
19 |
21 |
23 |
42 |
33 |
23 |
89 |
81 |
78 |
4 |
Enterobacter |
45 |
8 |
11 |
23 |
22 |
18 |
45 |
37 |
37 |
97 |
95 |
82 |
97 |
Citrobacter |
57 |
14 |
28 |
28 |
28 |
21 |
50 |
42 |
64 |
86 |
86 |
79 |
100 |
Acinetobacter |
15 |
- |
10 |
0 |
10 |
10 |
25 |
15 |
10 |
40 |
35 |
35 |
100 |
Serratia |
57 |
29 |
29 |
0 |
57 |
0 |
71 |
57 |
71 |
100 |
100 |
100 |
100 |
100% Staphylococcus aureus, CoNS and Enterococcus were sensitive to vancomycin. Staph aureus were also mostly sensitive to linezolid and teicoplanin was very effective antibiotic in vitro against Staphylococci (table IV).
Table IV: Antibiotic sensitivity pattern of gram positive bacteria isolated from wound swab and pus.
Organism |
Sensitivity (%) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Ak |
Aml |
Cro |
Cip |
Cot |
Do |
G |
L |
Lzd |
Fox |
CD |
Tec |
V |
|
Stahaureus |
41 |
- |
34 |
20 |
29 |
35 |
39 |
24 |
98 |
68 |
76 |
86 |
100 |
CoNS |
62 |
- |
62 |
31 |
38 |
62 |
69 |
38 |
100 |
62 |
69 |
69 |
100 |
Streptococcus |
50 |
88 |
100 |
50 |
50 |
50 |
75 |
63 |
- |
- |
- |
- |
- |
Enterococci |
0 |
50 |
- |
100 |
- |
50 |
0 |
50 |
100 |
- |
- |
- |
100 |
Discussion
Despite of application of the basic principles in wound care, a number of patients develop infections that require proper identification of the organisms for appropriate management. A changing pattern of isolated organism and their antimicrobial sensitivity which varies from hospital to hospital is a usual feature.
Development and spread of antibiotic resistance can be controlled by appropriate antimicrobial use, strict infection control, and continued surveillance.10
In the present study, about three fourth samples showed growth of bacteria on culture and it was comparable with growth rate reported in several other studies.17,18 In consistent with other studies,19,20 most of the isolated organisms from wound swab (90.4%) and pus (77.4%) were gram negative bacilli. In contrast to the present findings almost equal proportion of gram positive and gram negative bacteria were reported from pus and wound swab in some studies.20,21 Pattern of bacteria causing wound infection depends on hospital environment and surgical procedure involved.6
Among gram negative bacteria Pseudomonas spp was the most commonly isolated organism in this study followed by E. coli, Klebsiella spp and Proteus spp. In accordance, Pseudomonas spp was found to be the most common gram negative bacteria in a study in India.22 The frequency of Pseudomonas spp. as the causative agent of wound infection was 28% in 2011 and 26.5% in 2016 in Bangladesh, which is lower than this study.23,24 This higher rate of Pseudomonas infection might be due to the fact that majority of the wound swabs samples were sent from the burn unit of the hospital. Pseudomonas spp is a ubiquitous and versatile human opportunistic pathogen and from the endogenous gastrointestinal flora or environmental source it comprises one of the most common causative agents of burn wound infection.25,26 Pseudomonas produces both cell-associated and extracellular virulence factors that mediate a number of processes, including adhesion, leukocyte killing, tissue destruction, immune system evasion and bloodstream invasion that make it an efficient agent for infection in burn wound.27,28 In contrast to the present findings E. coli and Klebsiella spp were reported as the most predominant gram negative bacterial pathogen.18,19,29 The discrepancy of the isolation rate may be due to varying prevalence of infection causing bacteria from hospital to hospital as because different hospital deals with different types of infection.
In this study, Staphylococcus aureus was the most frequently isolated gram positive bacteria from both wound swab and pus which was similar to the other studies done in Bangladesh and India.17,18,30 About 32% of the isolated Staphylococcus aureus were Methicillin resistant Staphylococcus aureus (MRSA). In Bangladesh, the rate of MRSA infection ranges from 32% to 63% in different studies which is in accordance to the isolation rate in this study.31 MRSA is a multidrug resistant bacterium which is resistant to methicillin and other penicillins and most cephalosporins, â-lactam/â-lactamase inhibitor combinations and carbapenems.14
The antibiotic susceptibility data in this study showed that some common antibiotics have very limited usefulness for treatment of wound infections. Highest resistance by gram negative bacilli was noted against amoxicillin followed by fluoroquinolones, co- trimoxazole, and third generation cephalosporins in our study. This pattern of resistance has been shown by several studies.32,33 Ciprofloxacin is an important antibiotic but gram negative bacilli showed high resistance to it. This finding is consistent with studies who reported gram negative bacteria were highly resistant to ciprofloxacin.34-36 The higher rate of resistance to ciprofloxacin might be due to the fact that this drug is used widely in Bangladesh for many infections such as enteric fever that is endemic here. Majority of the gram negative bacilli showed a comparatively good sensitivity to amikacin and the pattern is consistent with other studies in Bangladesh.34-36 The reason behind such low resistance might be the less use of this antibiotic in this hospital. The result indicates that amikacin may be considered as an alternative drug in infections caused by gram negative bacilli in this setting.
Carbapenems were very effect antibiotics showing 66-97% sensitivity against gram negative bacilli except Acinetobacter spp in this study, which is in accordance with findings of several studies.34,35 Though only few cases showed resistance but this resistance to carbepenems is a matter of great concern in treatment of infection. Colistin and piperacillin/tazobactum showed higher rate of sensitivity in this study. All isolated Klebsiella and Acinetobacter spp were sensitive to colistin, and except for Pseudomonas and Acinetobacter spp piperacillin/tazobactum showed good sensitivity to gram negative bacilli. These two injectable drugs are usually considered as reserve drug and are being used for those who are resistant to most other antibiotics.
In this study, 14.6% of the gram negative bacteria were detected as ESBL producer which was in accordance with a study in 2017.37 In Bangladesh, the rate of ESBL producing bacteria were 23% in 2008 and 24% in 2012 which is higher than the present findings.35,38 Reduced use of cephalosporins and comparatively more use of carbapenems and colistin in treatment of infection by gram negative bacteria in present time in this hospital setting may be an important reason for reduced proportion of ESBL producers in the present study. Low rate of ESBL in this study could also be due to inclusion of all indoor and outdoor samples while previous studies were done on infected surgical wound only or ICU patients. E. coli and Klebsiella spp were the most commonly isolated ESBL producing organisms in this study. Detection of ESBL-mediated resistance is of great importance because of limited therapeutic options.15
Gram positive bacteria were mostly sensitive to vancomycin and linezolid. All the Staphylococcus aureus were sensitive to vancomycin, followed by linezolid (98%) and teicoplanin (86%); and had reduced sensitivity to ciprofloxacin (20%), levofloxacin (20%) and Co-trimoxazole (29%) which was similar to the sensitivity pattern of Staph aureus in a study in 2019 in Bangladesh.39
Overall antibiotic susceptibility data in this study shows that with few exceptions bacteria were found more resistant to antibiotics used in oral form. In Bangladesh, antibiotics especially oral antibiotics are common subjects of indiscriminant uses; antibiotics are sold over the counter in some places and anybody can purchase it even without prescription. Moreover, drug sellers often offer alternative oral antibiotics instead of the prescribed one. There is also lack of awareness among some patients about maintaining the prescribed right dose and duration of antibiotic. As a result of such factors antibiotic resistance is very common in Bangladesh.
Conclusion
There are very limited treatment options available for the resistant bacteria. So, early detection and appropriate antibiotic application remain a significant priority in controlling the development and spread of multidrug resistant organisms. The findings of this study provide an insight to the current state of causative organisms of wound swabs and pus and their sensitivity pattern. An effective national level antibiotic policy along with infection control measures should be introduced to preserve the effectiveness of antibiotics.
References
- Gottrup F, Melling A, Hollander D. An overview of surgical site infections: aetiology, incidence and risk factors. European Wound Management Association J. 2005; 5: 11-15.
- Isibor JO, Oseni A, Eyaufe A, Osagie R, Turay A. Incidence of aerobic bacteria and Candida albicans in post- operative wound infection. African Journal of Microbiology Research. 2008; 2: 288-91.
DOI: 10.5897/AJMR.9000407 - Cogen AL, Nizet V, Gallo RL. Skin microbiota: a source of disease or defense? Brit J Dermatol. 2008; 158: 442-55.
DOI: 10.1111/j.1365-2133.2008.08437.x - Dryden MS. Complicated skin and soft tissue infection. J Antimicrob Chemother. 2010; 65: 35-44.
DOI: 10.1093/jac/dkq302 - Rai S, Yadav UN, Pant ND, Yakha JK, Tripathi PP, Poudel A, et al. Bacteriological profile and antimicrobial susceptibility patterns of bacteria isolated from pus/wound swab samples from children attending a tertiary care hospital in Kathmandu, Nepal. Int J Microbiol. 2017; ID 2529085: 1-5
DOI: 10.1155/2017/2529085 - Mangram AJ, Horan TC, Pearson ML, Silver LC, Jarvis WR. Guideline for Prevention of Surgical Site Infection. Centers for Disease Control and Prevention (CDC) Hospital Infection Control Practices Advisory Committee. Am J Infect Control. 1999; 27: 97-132.
DOI: 10.1086/501620 - Mahmood A. Bacteriology of surgical site infections and antibiotic susceptibility pattern of the isolates at a tertiary care hospital in Karachi. J Pak Med Assoc. 2000; 50: 256- 59.
PMID: 10992708 - Cantlon CA, Stemper ME, Schwan WR, Hoffman MA, Qutaishat SS. Significant pathogens isolated from surgical site infections at a community hospital in the Midwest. Am J Infect Control. 2006; 34: 526-29.
DOI: 10.1016/j.ajic.2006.04.206 - Sharafati-chaleshtori R, Sharafati-chaleshtori F, Karimi A. Antibiotic resistance pattern of Staphylococcus strains isolated from orange and apple juices in Shahre-kord, Iran. Pak J Med Sci. 2010; 26: 615-18.
- Goswami NN, Trivedi HR, Goswami APP, Patel TK, Tripathi CB. Antibiotic sensitivity profile of bacterial pathogens in postoperative wound infections at a tertiary care hospital in Gujarat, India. J Pharmacol Pharmacother. 2011; 2: 158- 64.
DOI: 10.4103/0976-500X.83279 - Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG. Multi drug resistant, extensively drug resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18: 268-81.
DOI: 10.1111/j.1469-0691.2011.03570.x - Rice LB. Antimicrobial resistance in gram-positive bacteria. Am J Med. 2006; 119: 11-19.
DOI: 10.1016/j.amjmed.2006.03.012 - Iredell J, Brown J, Tagg K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. Brit Med J. 2016; 352: 64-72.
DOI: 10.1136/bmj.h6420 - CLSI. Performance standards for antimicrobial susceptibility testing; Twenty-third informational supplement, CLSI document M100-S25, Wayne, PA: Clinical and Laboratory Standards Institute, 2015.
- Paterson DL and Bonomo RA. Extended-spectrum beta- lactamases: a clinical update. Clin Microbiol Rev. 2005; 18: 657-86.
DOI: 10.1128/CMR.18.4.657-686.2005 - Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966; 36: 493-96.
- Anbumani N, Kalyani J, Mallika M. Epidemiology and microbiology of wound infections. Indian Journal for the Practicing Doctor. 2006; 3: 11-12.
- Tarana MN, Fardows J, Farhana N, Khatun R, Akter S. Bacteriological profile of wound swab and their antimicrobial susceptibility pattern in Shaheed Suhrawardy Medical College, Dhaka. Journal of Shaheed Suhrawardy Medical College. 2019; 11: 65-68.
DOI: 10.3329/jssmc.v11i1.43183 - Gangania PS, Singh VA, Ghimire SS. Bacterial isolation and their antibiotic susceptibility pattern from post-operative wound infected patients. Indian J Microbiol Res. 2015; 2: 231-35.
DOI: 10.5958/2394-5478.2015.00020.5 - Anusha S, Vijaya LD, Pallavi K, Manna PK, Mohanta GP, Manavalan R. An epidemiological study of surgical wound infections in a surgical unit of tertiary care teaching hospital. Indian Journal of Pharmacy Practice. 2010; 3: 8-13.
- Zafar A, Anwar N, Ejaz H. Bacteriology of infected wounds- A study conducted at children hospital Lahore. Biomed. 2008; 24: 71-74.
- Kumar R, Kumar A, Keshri UP, Gari M, Mahato SK, Protim P. Antimicrobial susceptibility pattern of pus culture in a tertiary care hospital of Jharkhand, India. Int J Basic Clin Pharmacol. 2017; 6: 1184-92.
DOI: 10.18203/2319-2003.ijbcp20171674 - Saha SK, Muazzam N, Begum SA, Chowdhury A, Islam MS, Parveen R. Study on time-related changes in aerobic bacterial pattern of burn wound infection. Faridpur Med College J. 2011; 6: 41-45.
DOI: 10.3329/fmcj.v6i1.7410 - Rahman M, Jobayer M, Akter N, Ahamed F, Shamsuzzaman SM, Mamun KZ. Rapid detection of Pseudomonas at species level by multiplex PCR in surgical units and ICU of Dhaka Medical College Hospital. Bangladesh J Med Microbiol. 2016; 10: 22-26.
- Franco BE, Martinez MA, Rodriguez MAS, Wertheimer AI. The determinants of the antibiotic resistance process. Infect Drug Resist. 2009; 2: 1-11.
PMCID: PMC3108730 - Altoparlak U, Erol S, Akcay MN, Celebi F, Kadanali A. The time-related changes of antimicrobial resistance patterns and predominant bacterial profiles of burn wounds and body flora of burned patients. Burns. 2004; 30: 660-64.
DOI: 10.1016/j.burns.2004.03.005 - Tredget EE, Shankowsky HA, Rennie R, Burrell RE, Logsetty S. Pseudomonas infections in the thermally injured patient. Burns. 2004; 30: 3-26.
DOI: 10.1016/j.burns.2003.08.007 - Van-Delden C and Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998; 4: 551-60.
DOI: 10.3201/eid0404.980405 - Nirmala S and Sengodan R. Aerobic bacterial isolates and their antibiotic susceptibility pattern from pus samples in a tertiary care government hospital in Tamilnadu, India. Int J Curr Microbiol App Sci. 2017; 6: 423-42.
DOI: 10.20546/ijcmas.2017.606.050 - Shriyan A, Sheetal R, Nayak N. Aerobic micro-organisms in post-operative wound infections and their antimicrobial susceptibility patterns. J Clin Diagn Res. 2010; 4: 3392-96.
- Haq JA, Rahman MM, Asna SM, Hossain MA, Ahmed I, Haq T, et al. Methcillin-resistant Staph aureus in Bangladesh: a multicentre study. Int J Antimicrobiol Agents. 2005; 25: 276-77.
DOI: 10.1016/j.ijantimicag.2005.01.004 - World Health Organization. Community-based surveillance of antimicrobial use and resistance in resource constrained settings. Report on ûve pilot projects. WHO/EMP/MAR/2009.
- Umadevi S, Kandhakumari G, Joseph NM, Kumar S, Easow JM, Stephen S, et al. Prevalence and antimicrobial susceptibility pattern of ESBL producing gram negative bacilli. J Clin Diagn Res. 2011; 5: 236-39.
- Begum S, Salam MA, Alam KF, Begum N, Hassan P, Haq JA. Detection of extended spectrum â-lactamase in Pseudomonas spp. isolated from two tertiary care hospitals in Bangladesh. BMC Res Notes. 2013; 6: 2-4.
Available from: biomedcentral.com/1756-0500/6/7 - Farzana R, Shamsuzzaman SM, Mamun KZ, Shears P. Antimicrobial susceptibility pattern of extended spectrum â-lactamase producing Gram-negative bacteria isolated from wound and urine in a tertiary care hospital, Dhaka city, Bangladesh. Southeast Asian J Trop Med Public Health. 2013; 44: 96-103.
PMID: 23682443 - Sarker JN, Bakar SMA, Barua R, Sultana H, Anwar S, Saleh AA, et al. Susceptibility pattern of extended spectrum â- lactamase (ESBL) producing Escherichia coli, Klebsiella spp. and Enterobacter spp. to Ciprofloxacin, Amikacin and Imipenem. J Sci Res Reports. 2015; 8: 1-9.
DOI: 10.9734/JSRR/2015/16933 - Jobayer M, Afroz Z, Nahar SS, Begum A, Begum SA, Shamsuzzaman SM. Antimicrobial susceptibility pattern of ESBL producing organisms isolated in a tertiary care hospital, Bangladesh. Int J Appl Basic Med Res. 2017; 7: 189-92.
DOI: 10.4103/ijabmr.IJABMR_28_16 - Islam MS, Yusuf MA, Begum SA, Sattar AFMA, Hossain A, Roy S. Extended spectrum beta-lactamase producing uropathogenic Escherichia coli infection in Dhaka, Bangladesh. J Bacteriol Res. 2015; 7: 1-7.
DOI: 10.3329/jsf.v10i2.17959 - Taz KA, Jobayer M, Shamsuzzaman SM. Nasal colonization of Methicillin resistant Staphylococcus aureus among healthcare providers in a tertiary care hospital, Bangladesh. Mymensingh Medical J. 2019; 28: 627-33.
PMID: 31391436
Department of Microbiology, National Center for Control of Rheumatic Fever & Heart Disease, Sher-E-Bangla Nagar, Dhaka, Bangladesh.
mjobayerk52@yahoo.com
 0000-0002-2808-4774
0000-0002-2808-4774
Submission
19 September 2020
Accepted
30 July 2021
Published
01 August 2021
Apply citation style format of Bangladesh Medical Research Council
Issue
Vol 47 No 2 (2021)
Section
Research Articles
Ethical Clearance
IRB of Dhaka Medical College, Dhaka, Bangladesh.
Financial Support
Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh.
Conflict of Interest
We do not have any potential conflicts of interest.